Review Diagnostic Discovery Results¶
Potential pathogenic variants identified through research or internal work at Genomics England are returned to Genomic Laboratory Hubs (GLHs) for clinical review via the Diagnostic Discovery pathway. Updating the reported outcomes for Diagnostic Discovery findings facilitates data population into the Clinical Variant Ark (CVA), informs the Known Pathogenic Variant Prioritisation (KPVP) tiering algorithm, and updates the Trusted Research Environment (TRE).
How to Update Cases¶
Video Tutorial
Video Tutorial¶
The following video provides an overview of the process that users can follow to update these cases.
Step 1. Identify Case in the Interpretation Portal
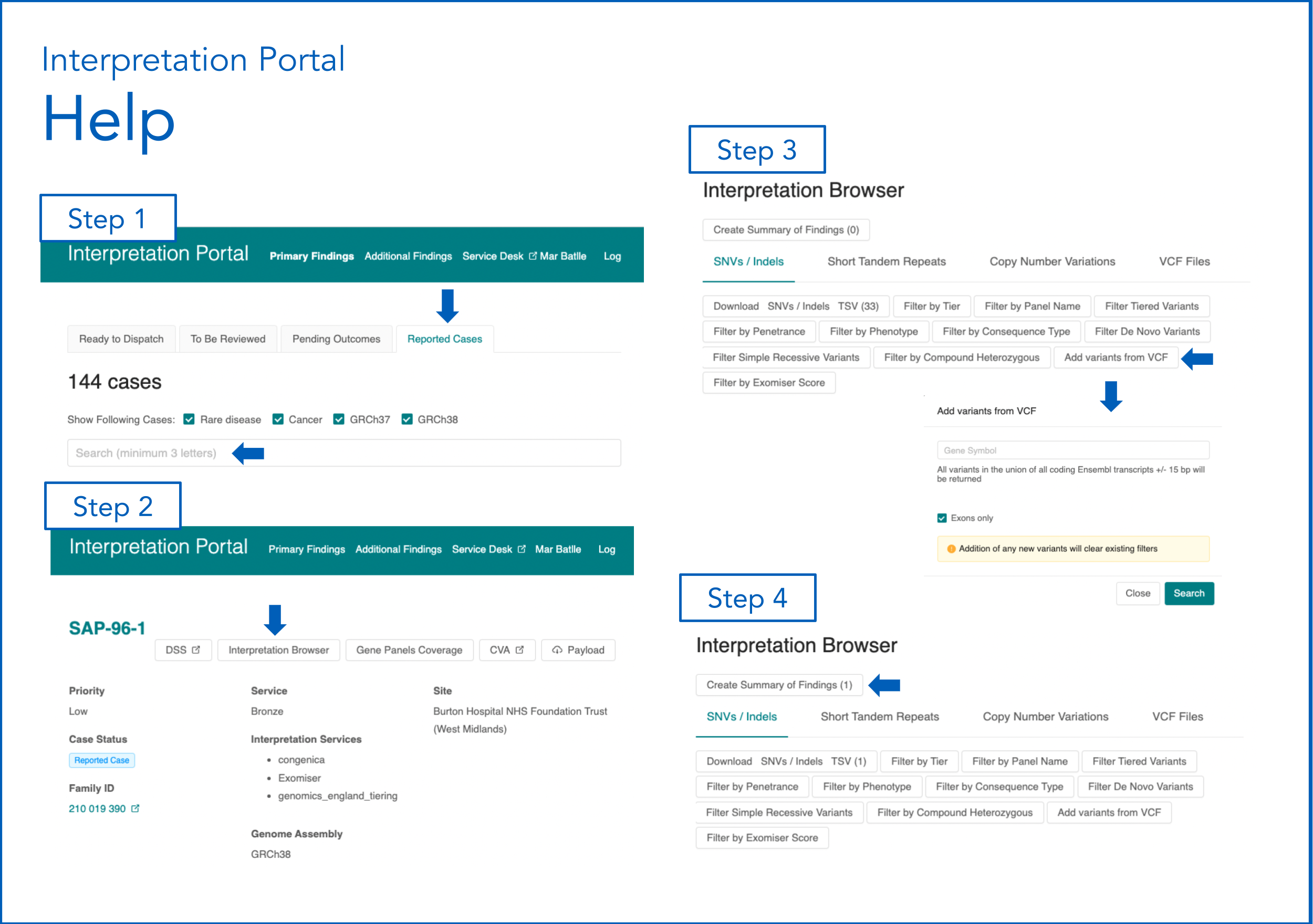
Step 1. Identify Case in the Interpretation Portal¶
- Log in to the Interpretation Portal:
GMS Interpretation Portal 100K Interpretation Portal
- Navigate to the 'Reported Cases' tab and search by participant ID.
- For family members outside the proband, search in CVA using the participant ID, then use the 'View on Interpretation Portal' link.
Step 2. Check if variant(s) is/are identifiable in the Interpretation Portal
Step 2. Check if variant(s) is/are identifiable in the Interpretation Portal¶
Small Variants¶
- In the Interpretation Portal, click 'Interpretation Browser'.
- Check if the variant is listed.
- If not found, use 'Add variants from VCF':
- Enter the gene symbol.
- Deselect 'Exons only'.
- Click 'Search'.
-
If still not found, check for alternative gene symbols on the HGNC website.
-
For GRCh37 referrals, due to the Cellbase v5 upgrade,
Add Variants from VCFmay be unavailable. Proceed to Step 4 if variant remains unidentifiable.
CNVs¶
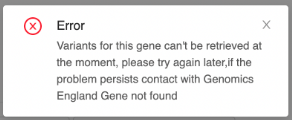
- For GRCh37 100k referrals or if the following warning appears, CNVs are unavailable; proceed to Step 4.

- In the Interpretation Portal, click 'Interpretation Browser' -> 'Copy Number Variations'.
- Search for the variant. If not found, proceed to Step 4.
STRs¶
- For GRCh37 100k referrals or if the following warning appears, STRs are unavailable; proceed to Step 4.

- In the Interpretation Portal, click 'Interpretation Browser' -> 'Short Tandem Repeats'.
- Search for the variant. If not found, proceed to Step 4.
Step 3. Variant(s) is/are identifiable in the Interpretation Portal
Step 3. Variant(s) is/are identifiable in the Interpretation Portal¶
- Click '+' to expand variant details.
- Select a report event, ensuring the correct mode of inheritance.
- For untiered/unranked Exomiser variants, select the appropriate mode of inheritance.
- Confirm no patient identifiable information and 'Save draft'.
- Repeat for other variants.
Step 4. Variant(s) is/are not identifiable in the Interpretation Portal
Step 4. Variant(s) is/are not identifiable in the Interpretation Portal¶
- Click 'Create Summary of Findings'.
- Add the variant description and variant consequence to the 'Your Interpretation' section, indicating that the variant couldn't be added. Example:
Couldn't add: 16:8347473957:T:A - missense - 'Save draft'.
- Repeat for other variants.
Step 5. Create Summary of Findings and Exit Questionnaire
Step 5. Create Summary of Findings and Exit Questionnaire¶
- Add previous findings.
- Click
Create Summary of Findingsto generate a report. - Complete the ‘Reporting Outcomes Questionnaire’.
- For manually added variants (Step 4), in ‘Rare Disease Reporting Outcomes’:
- "Have the results reported here explained the genetic basis of the family’s presenting phenotype(s)?": Yes
- "Additional Comments": "Add the variant details (chrom:pos:reference:alternate) and associated service desk ticket if applicable."