Frequently Asked Questions¶
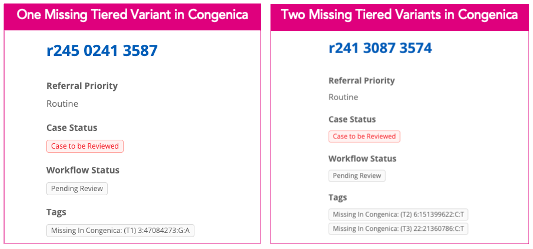
What are tags?
Tags in the Interpretation Portal alert users to specific issues impacting analysis. Managed by the CIP-API or bio-operations, they highlight deviations or potential concerns, such as the unavailability of specific variants in the DSS.
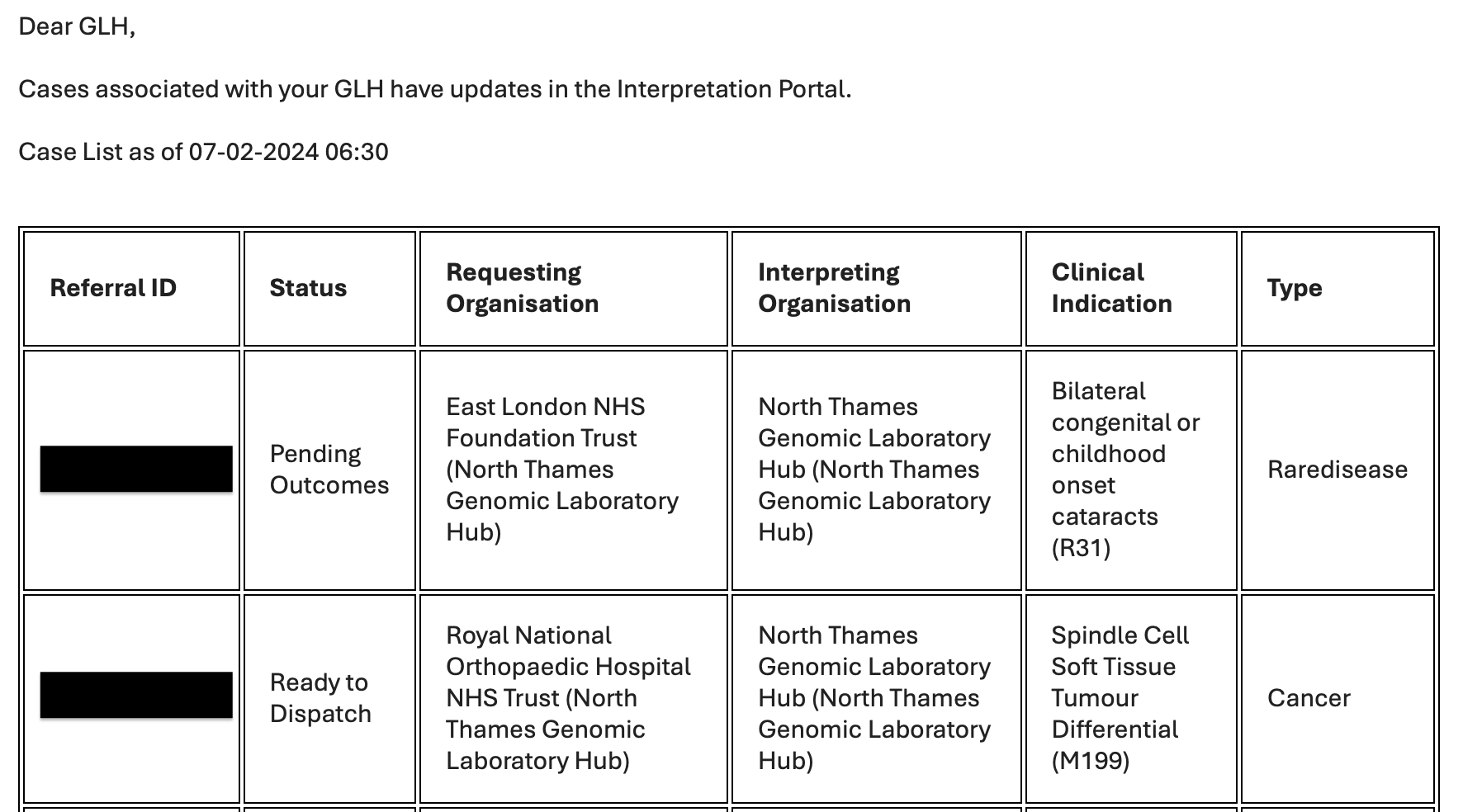
What are the email notifications?
Email Notifications provide daily alerts to GLHs about cases with status changes requiring attention. Separate emails are sent for routine and urgent referrals, each containing a table with key case details: Referral ID, Status, Requesting/Interpreting Organizations, Clinical Indication, and Type.
To update recipient emails, submit a Service Desk ticket.
What do the warnings or error messages mean?
Error/warning messages don't always indicate system errors.
- N/A under “NHS ID”, “Full Name”, “Date of Birth”, and “Patient Choice” means the portal can't connect to TOMS, which is expected when data is unavailable.
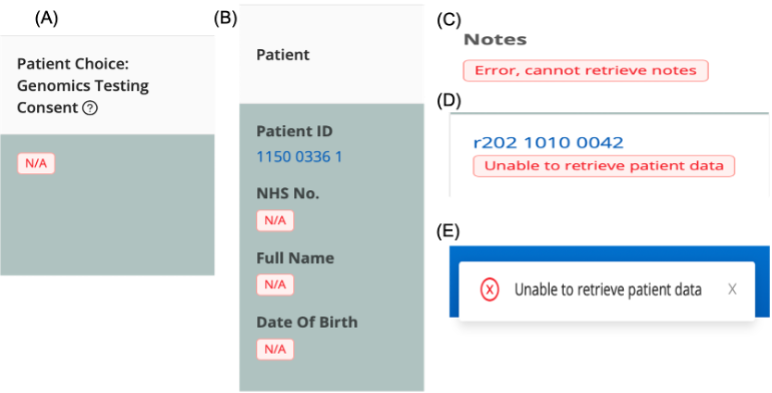
- Message A: Genomics Testing Consent cannot be populated from TOMS.
- Message B: Portal can't connect to TOMS to populate the case with PID.
- Message C: Portal can't connect to TOMS to populate case notes.
- Message D: Portal can't connect to TOMS to populate the case with PID.
- Message E: Portal can't connect to TOMS to populate the case with PID.
In the Referral Grid, "Unable to retrieve patient data" means the portal can't connect to TOMS to populate the case with PID.
What are Interpretation Services?
Interpretation Services are applications/algorithms that help determine the molecular basis of patient conditions. Each service produces an Interpreted Genome, a prioritized list of variants.
The Rare Disease Interpretation Browser allows review of these results and selection of primary findings.
For details on Tiering and Exomiser, see the Genomics England Rare Disease Genome Analysis Guide v2.0.
Note: An Exomiser score of 0 may make a variant appear missing, but it will have an Exomiser ranking. Review variants with ranks 1-3 and scores above 0.75. Check the Exomiser score in the case payload.
Why can't I see the Genomic and Data checks on the Rare Disease Referral page?
The WGS pipeline performs a QC check of reported sex against genetic sex. This check must pass for the pipeline to run, so the information was deemed unnecessary and removed.
What are Decision Support Systems?
Decision Support Systems (DSS) enable review and reporting of cases, facilitating MDTs and allowing exploration beyond Interpretation Service highlights. Refer to application-specific DSS user guides for details.
What is CVA?
The Clinical Variant Ark (CVA) is Genomics England's knowledge base for comparing cases with similar profiles. It offers a detailed overview of prioritized variants and includes linked SoFs, Reported Outcomes Questionnaires, and Variant Interpretation Logs.
Access CVA: CVA Portal.
What is CellBase?
CellBase supports annotation of genomic features by integrating biological information. The Interpretation Browser uses CellBase for additional variant annotations.
Why is my referral flagged?
If during processing, a case triggers a pre-defined QC check, they are automatically flagged. The table below lists the flags, a brief description and where they can be seen in the Interpretation Portal.
See the Referral Flags page for more information.
What Exomiser score is shown in the report events table?
The Interpretation Browser displays the "combined" Exomiser score. Individual variant and phenotype scores are in the payload. See the Exomiser Service Documentation.
Why is read depth information not shown for my variant?
Read depth information is displayed as reported by interpretation services. For homozygous reference variants, read depth appears as '-/-' within the Interpretation Browser. Note that read depth may differ from IGV due to filtering by variant calling and tiering.
When no read depth is reported, "N/A" is shown. Refer to IGV for read depth at these positions. Currently, read depth is not reported by Exomiser or for variants added from VCF for 100K referrals.
How does the Exomiser filter work?
The Exomiser filter is a "greater than" filter. It defaults to filtering variants with a combined score greater than 0.75.
What are the CVA classifications?
CVA classifications summarize classifications from reporting outcomes questionnaires. They don't include classifications added in DSS that are not in SoFs. These are in the Variant Interpretation Logs accessible via the CVA Portal.
What does the shading in the Interpretation Portal mean?
- Yellow-shaded referrals: Indicate draft work saved.
- Green-shaded variants: Indicate variants added from VCF or called by Exomiser but not yet validated.
How do I update a previous interpretation with new findings?
See here.