Upgrade to Cellbase v5¶
In the Nembus release, all NGIS applications are upgrading to Cellbase v5, our annotation system. Cellbase v5 introduces new and improved annotation datasets like Ensembl, UniProt, and ClinVar.
The resulting upgrades in the Interpretation Portal aim to enhance functionality and data availability, ensuring a smooth transition for users.
Changes Impacting Portal Features:¶
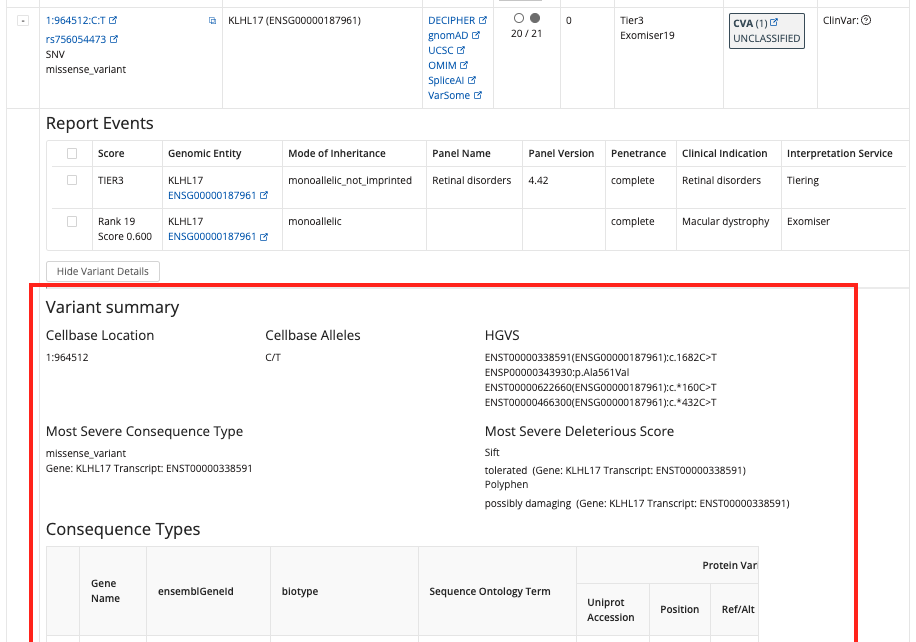
Show Variant Details¶
All information in this section now comes from Cellbase v5, including:
- CellBase Location
- Cellbase Alleles
- HGVS
- Most Severe Consequence Type
- Most Severe Deleterious Score
- Consequence Types
- Population frequencies
- Variant Trait Association
Users may see information differences if their case was processed with Cellbase version < 5.
This section should still load if a variant was retrieved using the Add Variants from VCF option.
GRCh37 100K cases
The Show Variant Details button is no longer available for GRCh37 cases.
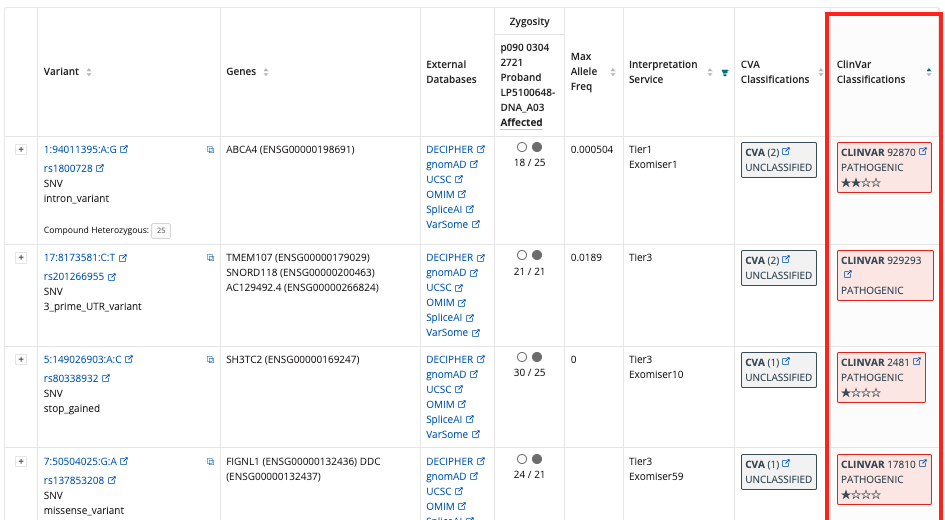
ClinVar Classifications¶
The Interpretation Browser uses Cellbase for the latest ClinVar Annotations. After this upgrade, users can continue using this feature as expected.
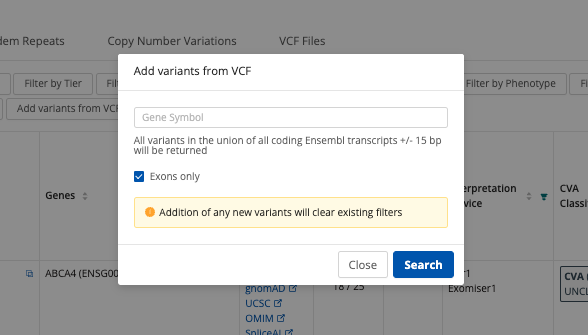
Add Variants from VCF¶
The Add Variants from VCF feature remains functional and will now retrieve the gene symbol using the latest Cellbase databases. If a case was processed with a version < 5, a different gene symbol than expected might appear due to the Portal using the most up-to-date information. This feature is no longer available for GRCh37 cases.
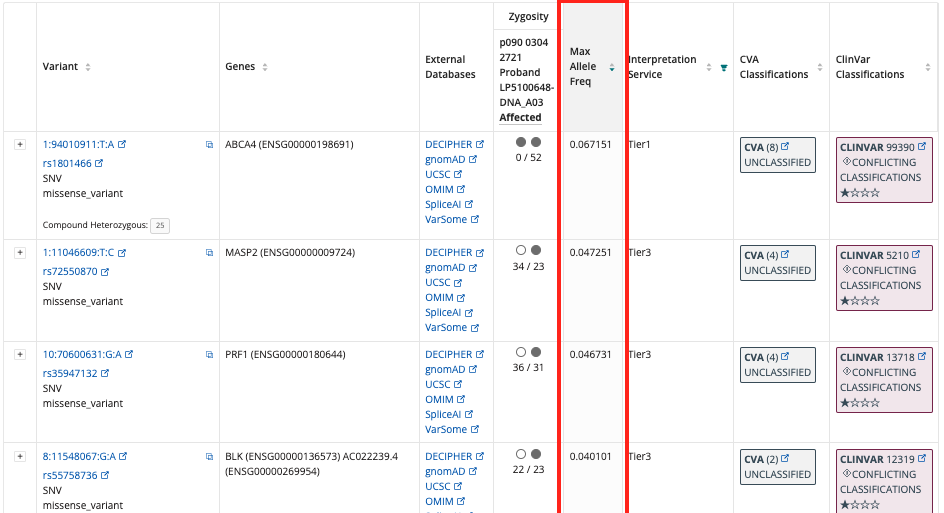
Max Allele Frequencies¶
The Interpretation Portal calculates Max Allele Frequencies using the current list of Studies and Subpopulations used by the Rare Disease Pipelines. With the Cellbase v5 upgrade, DiscovEHR and UK10K studies have been removed. Users will see the most up-to-date selection of studies, even if cases were processed with a Cellbase version < 5.
Summary of Findings (SoF)¶
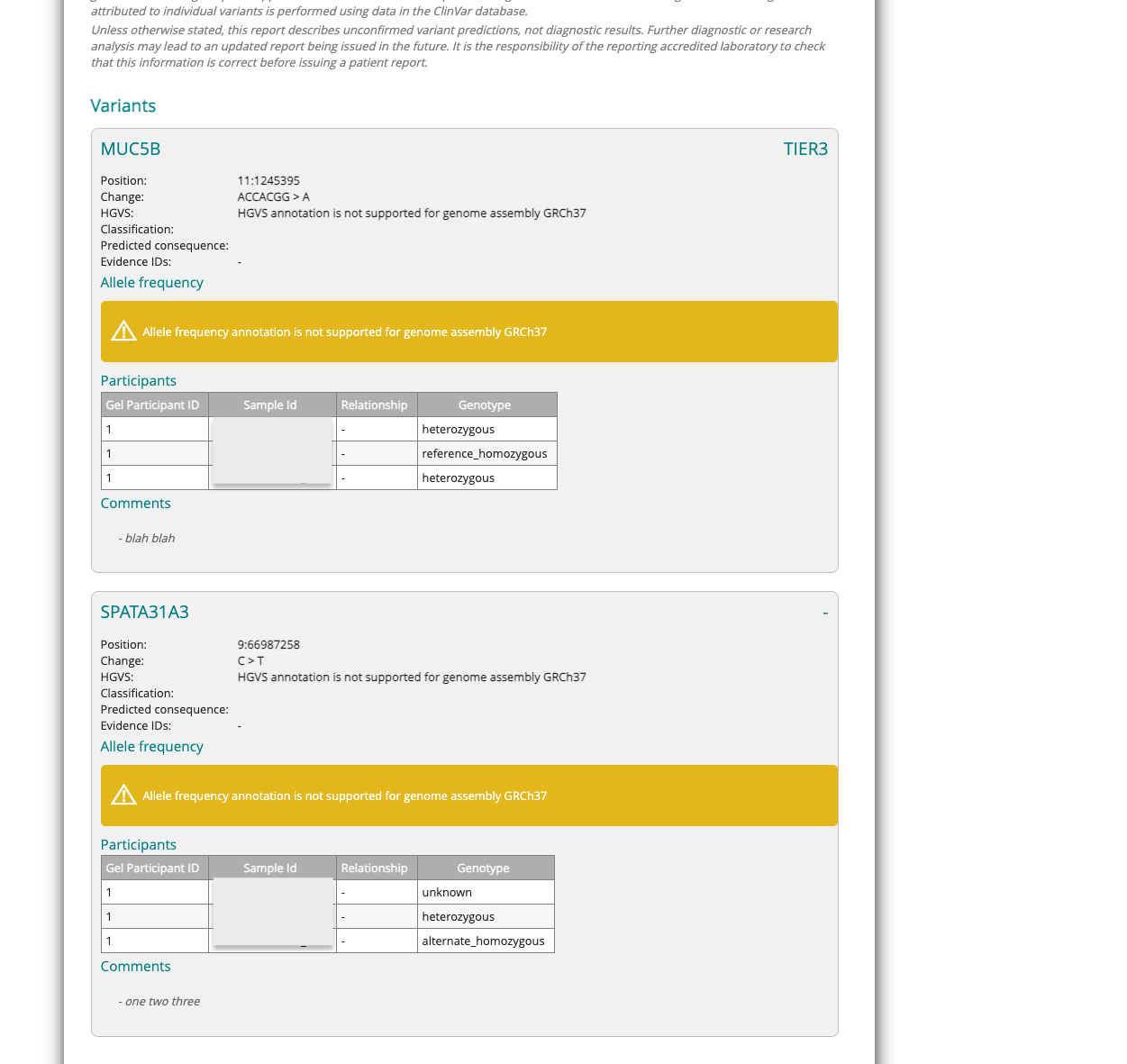
The Interpretation Portal uses Cellbase for Allele Frequency data when generating SoFs. This data will no longer be accessible for GRCh37 genome reference cases. Users will receive an error notification but can still download the SoF as usual.
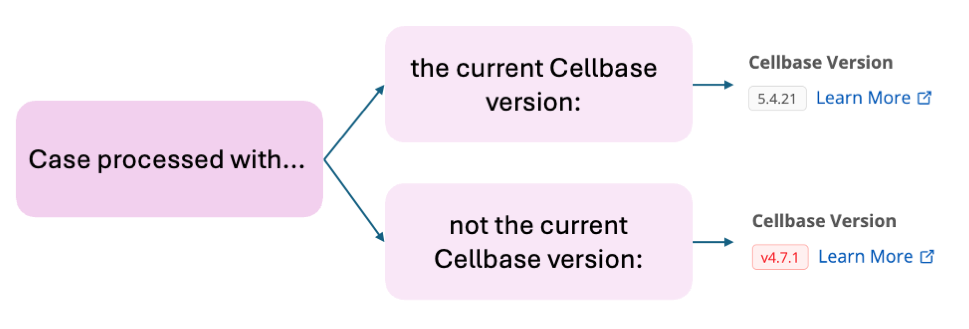
Identifying Cases Processed with Previous Cellbase Versions¶
Cases processed by the Genomics England Tiering pipeline before the Nembus release used an older Cellbase version. A new flag in the case information section of the Portal helps identify the Cellbase version used:
- Older version: The version number is highlighted in red.
- Current version: The version number is displayed without highlighting.
All cases will also include a link to this documentation page for further reference.
This flagging system allows quick identification of the Cellbase version used for processing and provides access to detailed information.