RD Interpretation Browser¶
The Interpretation Browser allows users to review Small Variants, Copy Number Variants (CNVs), Short Tandem Repeats (STRs), and generate a Summary of Findings (SoF) without requiring a Decision Support System (DSS). This tool also provides options for downloading variants in TSV format (with the ability to select specific variants) and related VCF files, which can be accessed from the VCF Files tab.
Reviewing Small Variants¶
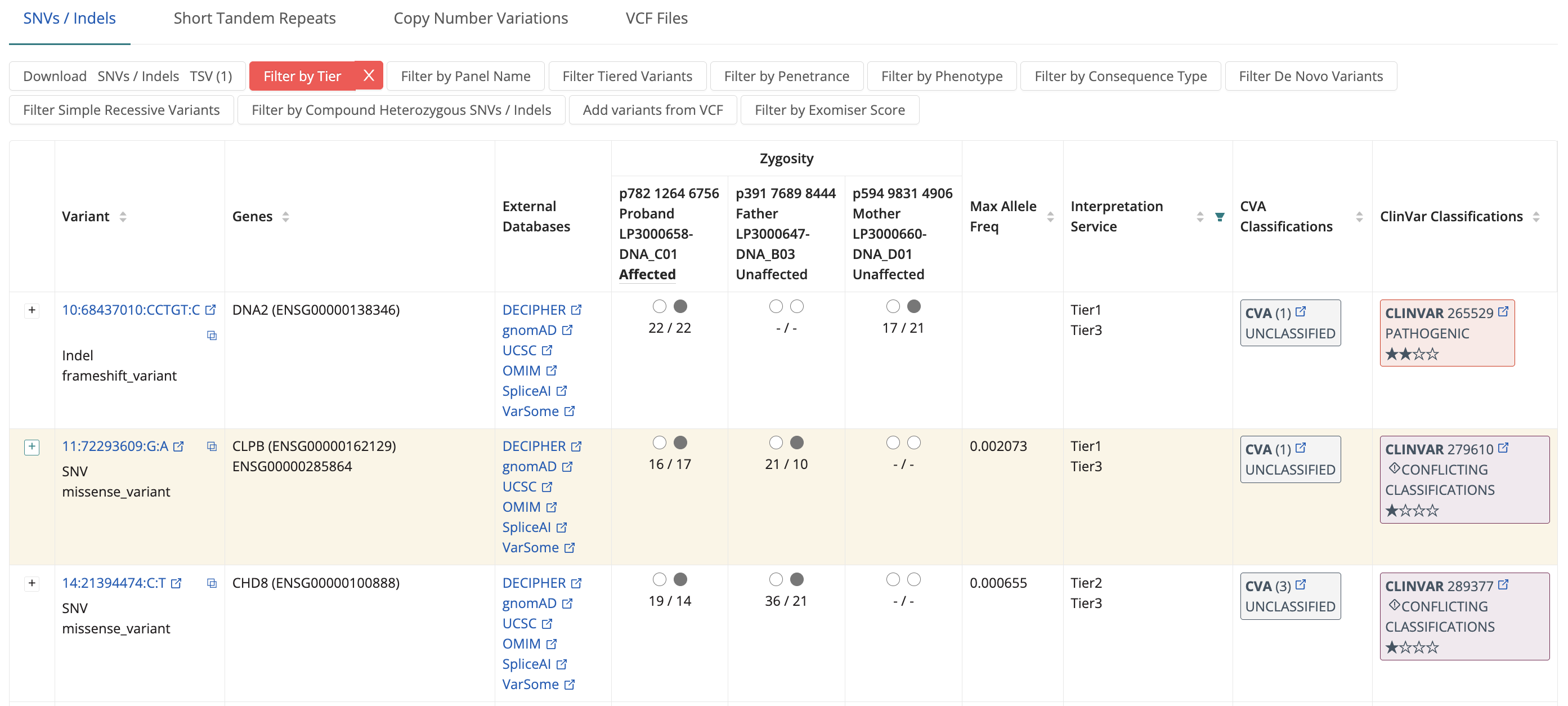
The SNV / Indels tab displays tiering results for small variants from the Genomics England Tiering and Exomiser.
Filter and Sort Small Variants
You can filter variants by: Tier and Exomiser Score, Panel Name, Penetrance, Phenotype, Consequence type, and Compound heterozygous group. You can also sort the variants by Positional details, Genes, Maximum Allele Frequency, CVA Classification, and Clinvar Classifications. This helps narrow down to the most relevant variants.
Why are the variants shaded?
Green indicates variants added from the VCF file or called by Exomiser but not yet validated, while orange indicates variants against which a decision has already been taken within this referral.
Add Variants from VCF
The Add Variants from VCF feature allows assessment of variants from genes outside applied panels. Enter the HGNC gene symbol and click Search to add PASS status variants for review.
- Variants must have
PASSstatus and a phenotype assigned to be added to the Summary of Findings. - This allows exploration of variants in genes not initially considered.
Only PASS status variants can be added to the Interpretation Browser. Non-PASS variants are viewable only in the Congenica DSS.
Review Variants
Once filtered, users can review variants. Clicking the 'reflected square' next to the coordinates copies variant details in Alamut format. Clicking variant coordinates allows checking read-level support using IGV.js. Linkouts to external databases (DECIPHER, gnomAD, UCSC, OMIM, SpliceAI, VarSome, CVA, ClinVar) provide further information. Zygosity across family members, indicated as Filled Black Circle (Presence), Single Black Circle (Heterozygosity), Two Black Circles (Homozygosity), and Dash ("-") (Undetermined), is crucial.
Clicking the + expands a variant to show report events, indicating scoring and evaluation against clinical indications. Information includes: Score, Genomic Entity, Mode of Inheritance, Panel Name, Panel Version, Penetrance, Clinical Indication, and Interpretation Service.
Clicking Show Variant Details retrieves additional annotations from Cellbase: Positional and allelic details, Most Severe Consequence Type, Consequence Types, Population Frequencies, Variant Trait Association, and Gene Trait Associated.
Reviewing Copy Number Variants¶
The Copy Number Variations tab displays tiering results for CNVs from the Genomics England Tiering.
B-Allele Frequency Graphs
After clicking on the CNV tab, the B-Allele Frequency Graphs will appear. Cases processed through the Genomics England RD pipeline generate B-Allele Frequency (BAF) graphs, downloadable in the CNV section. These graphs help detect potential mosaic CNV events, and each graph is directly downloadable as a file. For more information, see here in the Genomics England RD pipeline user guide. Below is an example of how the graphs are presented: 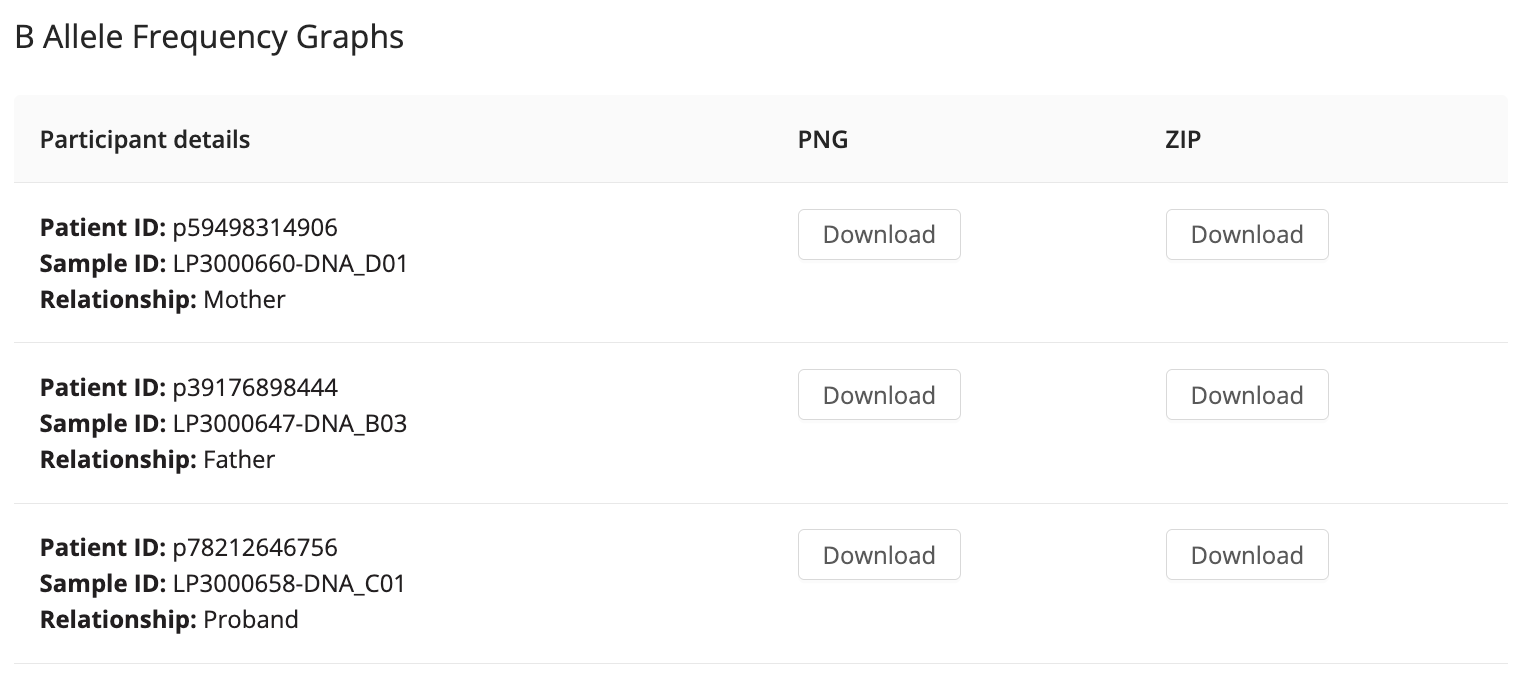
CNV tiering results are in two tables: “CNV Call Level Table” and “CNV Gene Level Table”.
CNV Call Level Table
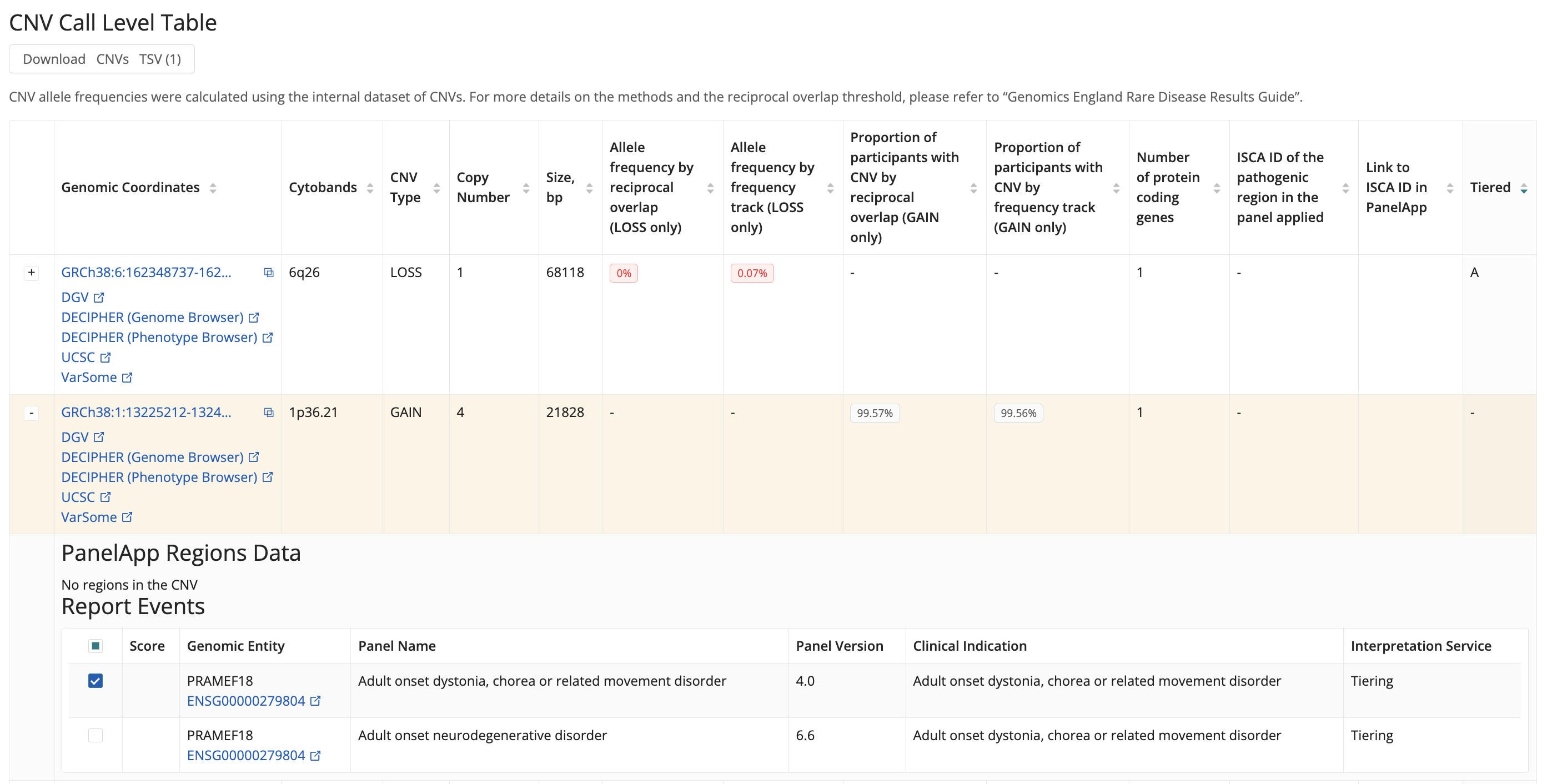
Lists all CNVs in the proband. Information includes: Genomic Coordinates (links to IGV, DGV, DECIPHER's Genome and Phenotype Browser, UCSC Genome Browser and VarSome), Cytobands, Type of CNV, Copy Number and Size, Allele Frequency by Reciprocal Overlap (LOSS only), Allele Frequency by Frequency Track (LOSS only), Proportion of Patients with CNV by Reciprocal Overlap (GAIN only), Proportion of Patients with CNV by Frequency Track (GAIN only), Number of Protein Coding Genes, ISCA ID of the Pathogenic Region in the Panel Applied, and Link to ISCA ID in PanelApp. Clicking the copy button in the variant column copies the variant details in Alamut format.
CNV Gene Level Table
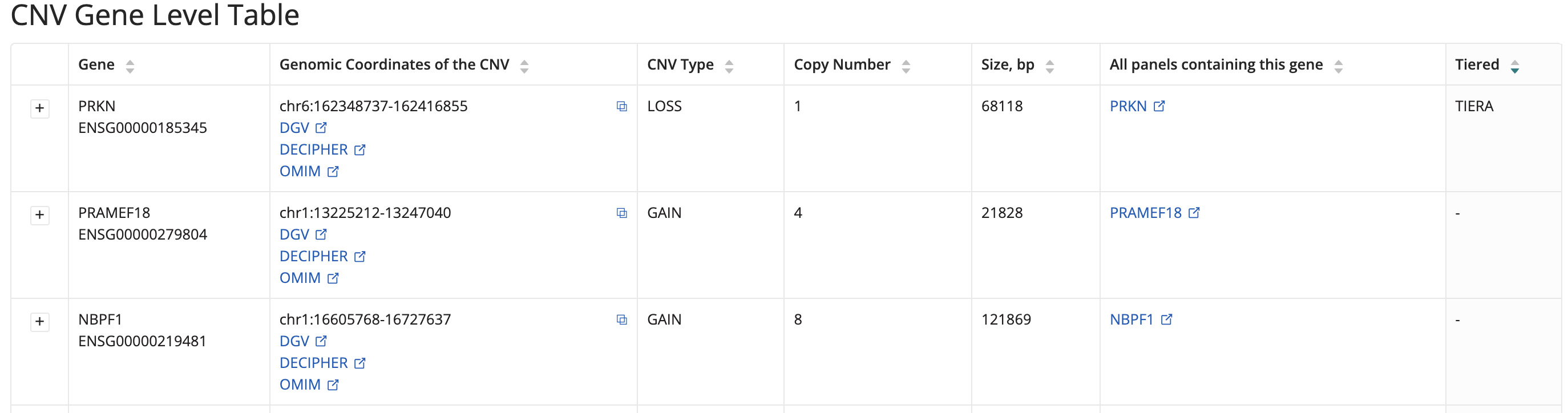
Lists genes overlapping CNV calls in the proband. Information includes: Gene (name and ENSG IDs), Genomic Coordinates of the CNV, CNV Type, Copy Number and Size, All Panels Containing this Gene (links to PanelApp), Tiered (TIERA or Null), and DGV link.
Reviewing Short Tandem Repeats¶
The Short Tandem Repeats tab displays tiering results for STRs from the Genomics England Tiering.
Filter and Review STRs
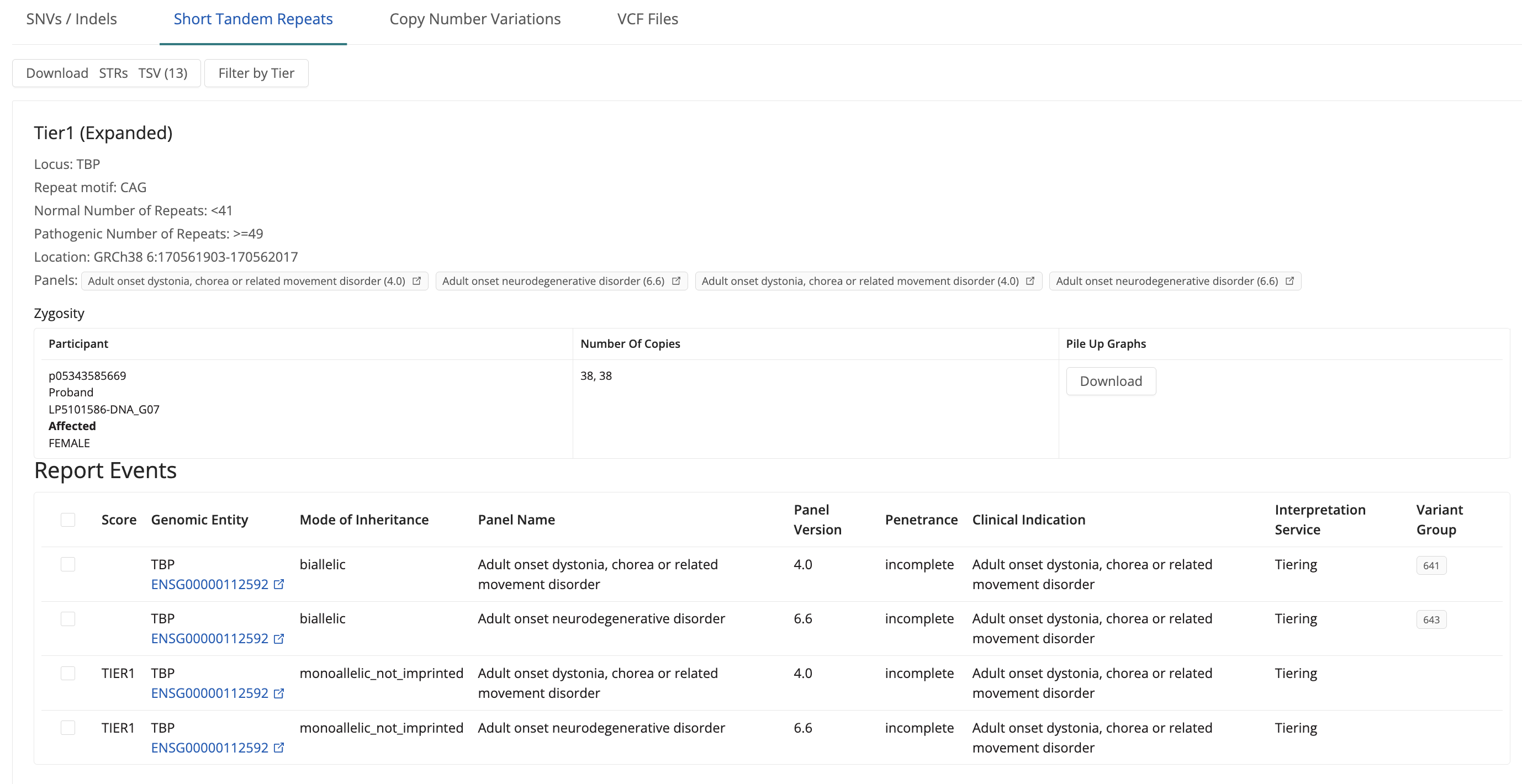
STRs are listed in priority order (Tier 1 > Tier 2 > Tier Null) and can be filtered based on the tier assigned. It displays information for each tiered variant: Tier, locus, repeat motif, repeat numbers, Genomic entity, Genomic coordinates, Panels applied to the case, Zygosity, Pile-Up Graph (download button), Mode of Inheritance, Panel name and version, and Penetrance of the clinical indication.
Generating a SoF without a DSS¶
A quick guide on how to close RD referrals without using a DSS.
Other Considerations¶
Unified Tiering¶
The RD pipelines use unified tiering for CNVs, STRs, and small variants, enabling the Interpretation Portal to show how different variant types impact a gene.
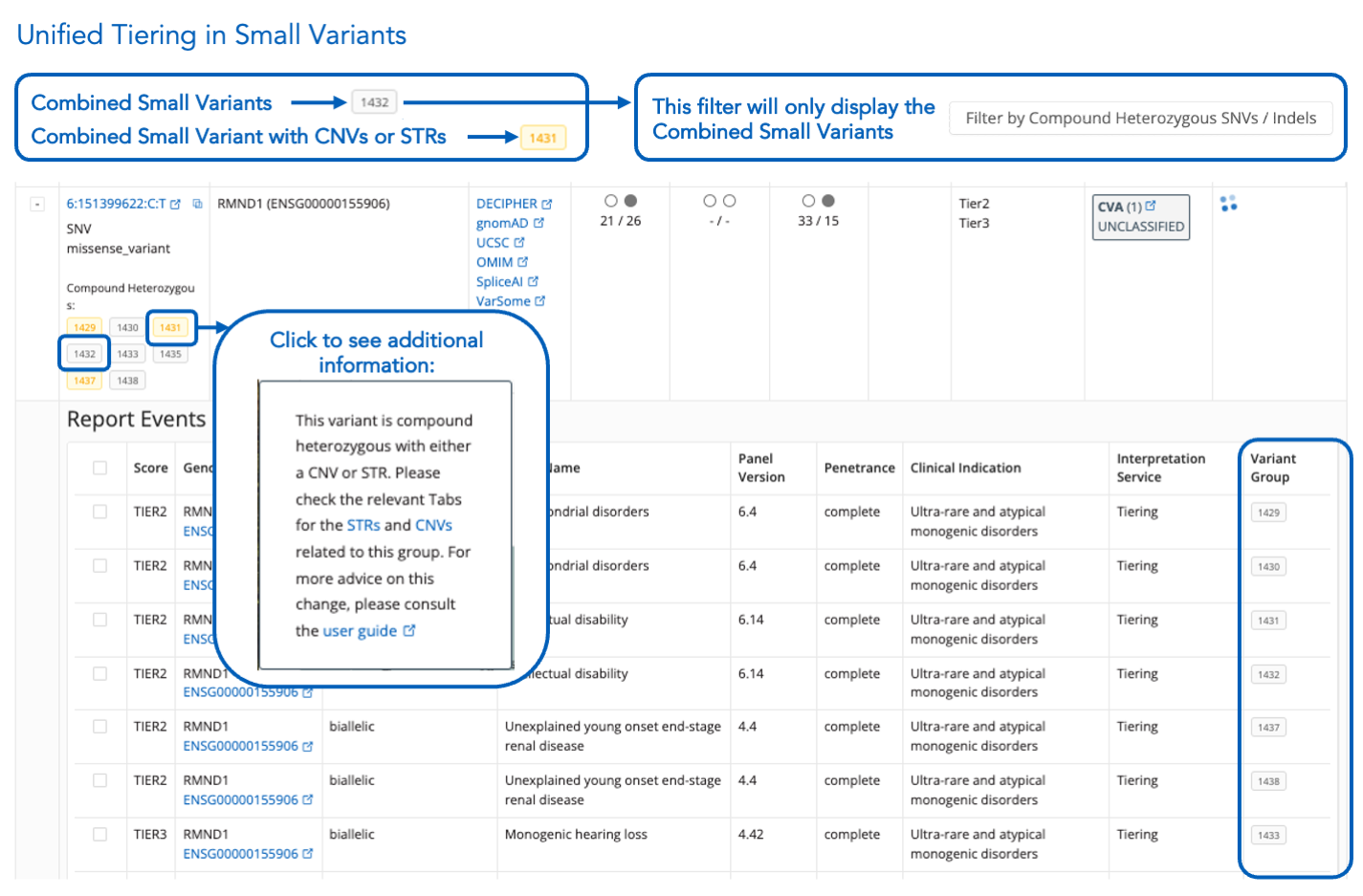
Unified Tiering for Small Variants
- Compound heterozygous variants are grouped in the "Variant" column (gray highlight). Use ‘Filter by Compound Heterozygous SNVs/Indels’ to locate them. The "Variant Group" column (visible when expanded) shows these groups.
- Variant groups combining small variants + CNVs/STRs are highlighted in yellow to differentiate them from small-variant-only groups. Clicking on these provides additional details, including a link to this guide.
- These groups won't appear when using the 'Filter by Compound Heterozygous SNVs/Indels’ option.
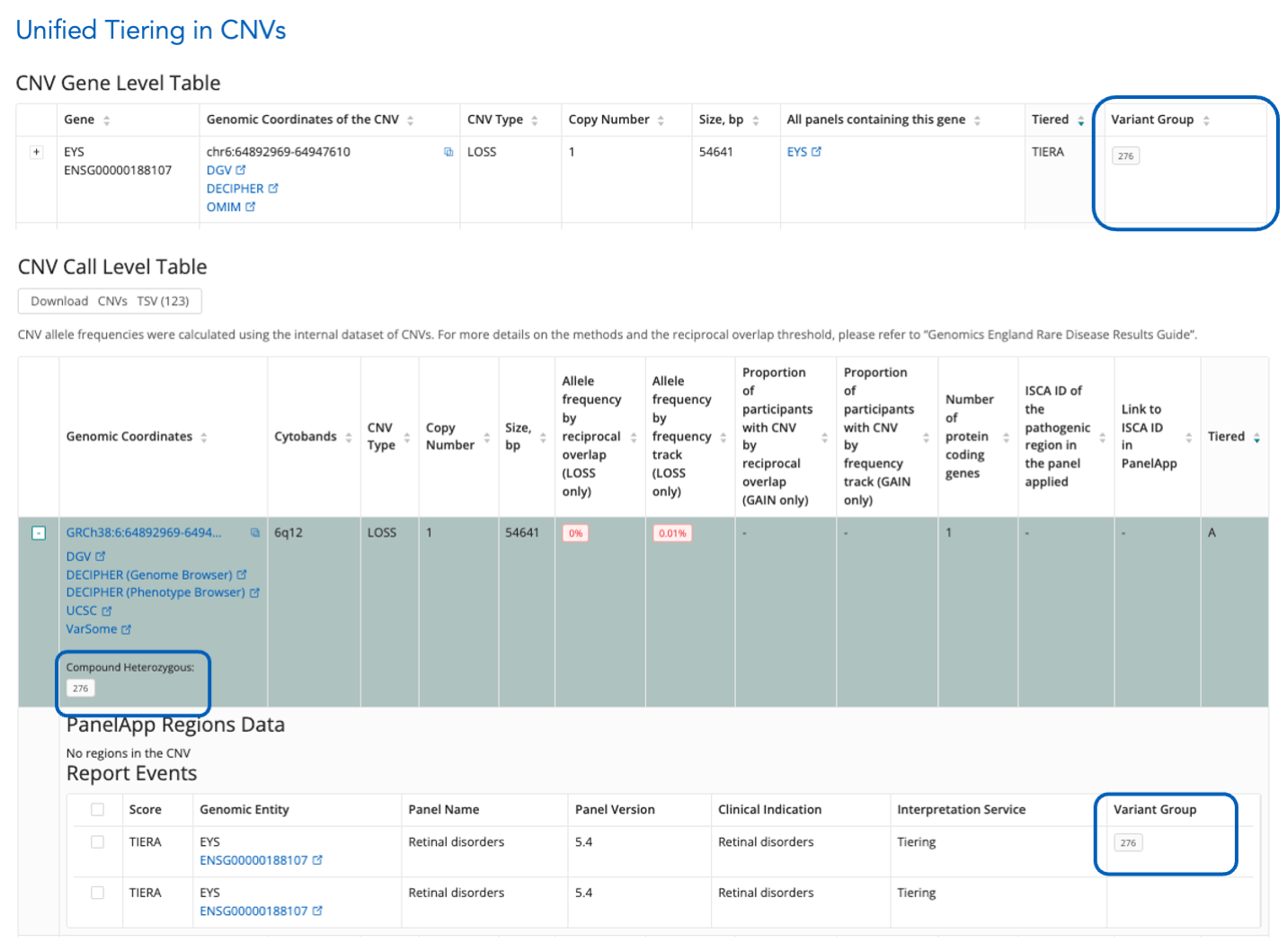
Unified Tiering for CNVs
- Call Level Table: Expand a CNV to view its group in the "Variant Group" column within the Report Events Table.
- Gene Level Table: The sortable "Variant Group" column shows compound heterozygous variantGroup values.
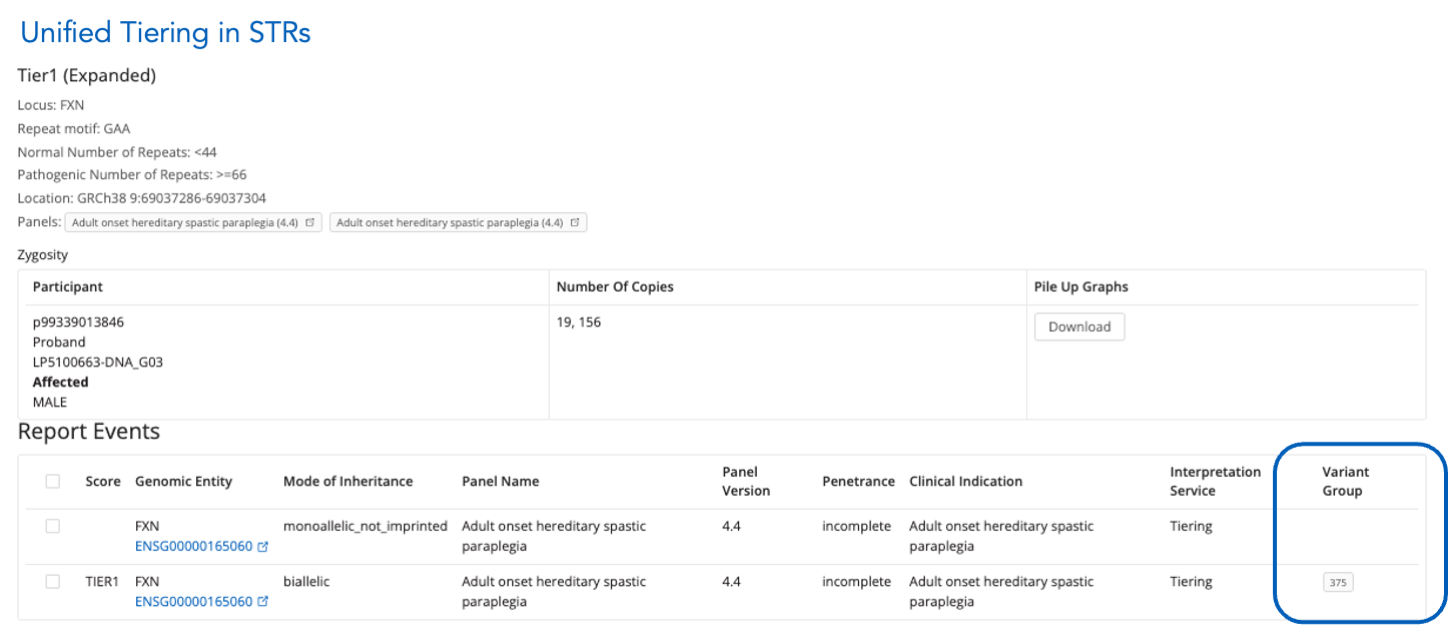
Unified Tiering for STRs
- If an STR is linked to a small variant, its group appears in the main view.
- Expanding the STR reveals the "Variant Group" column when tagged as Compound Heterozygous.